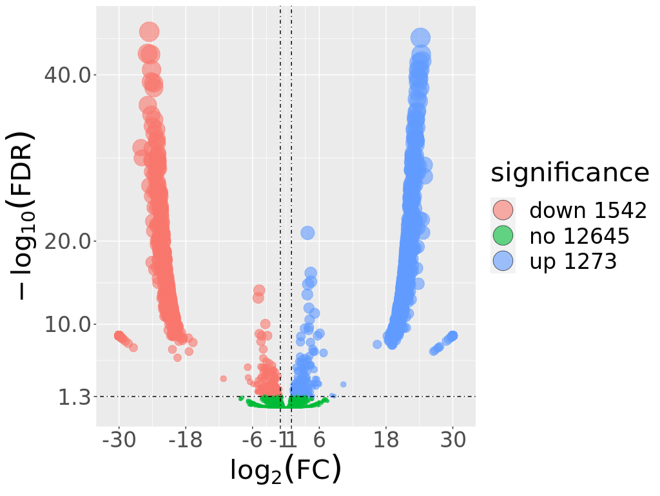
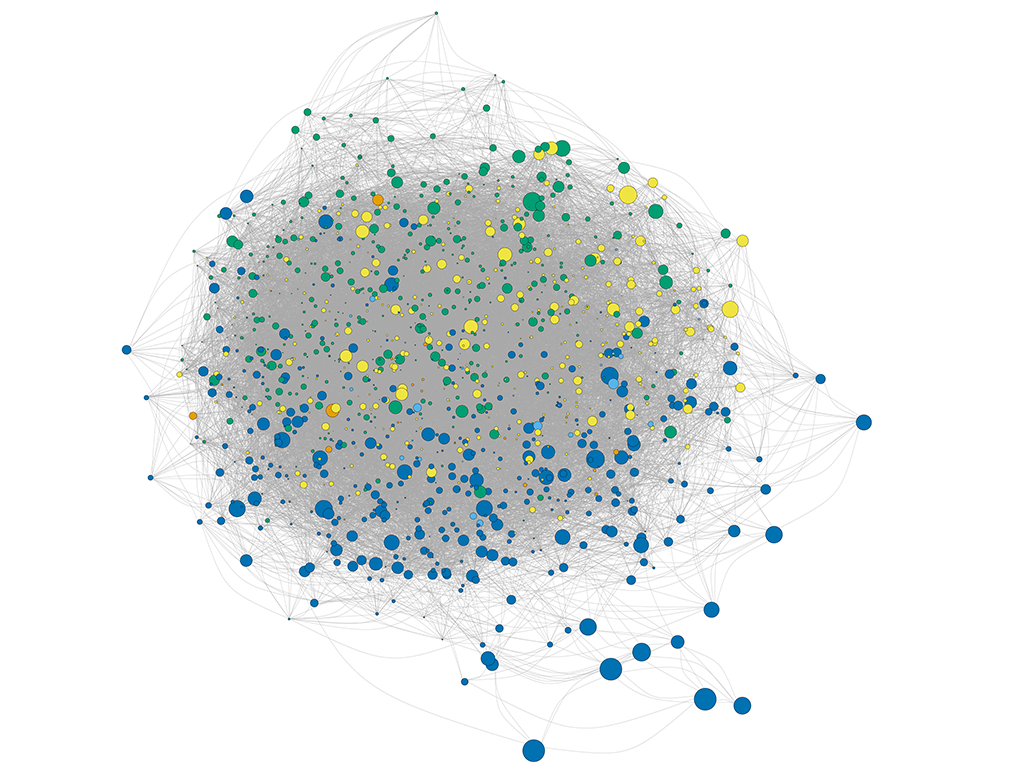
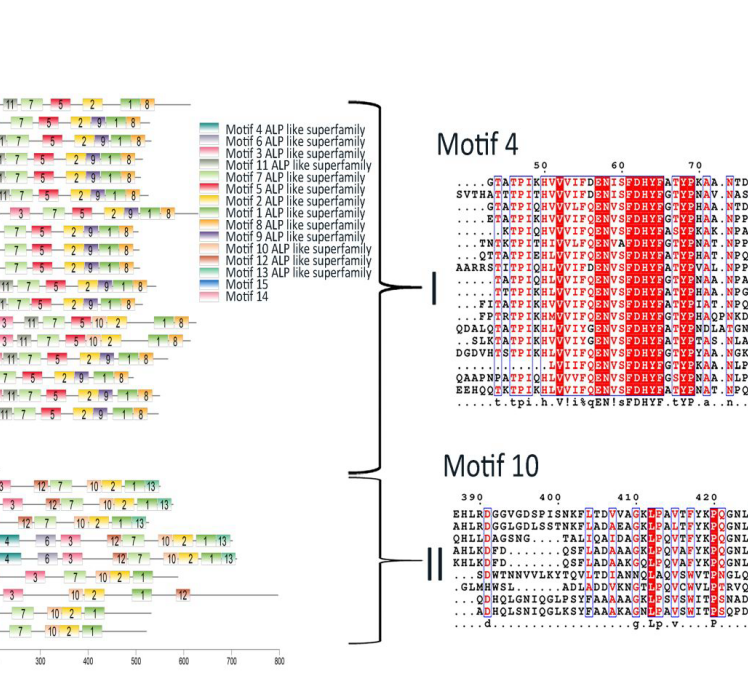
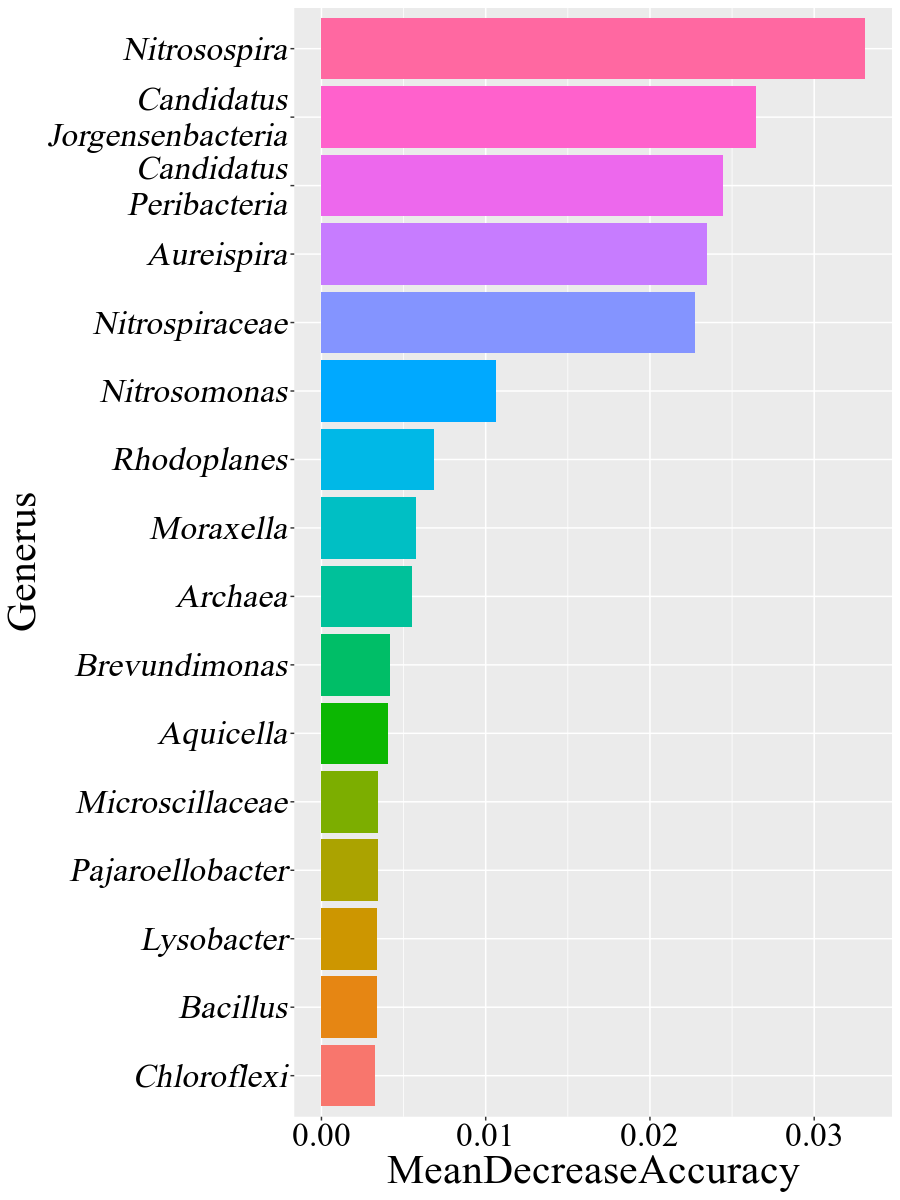
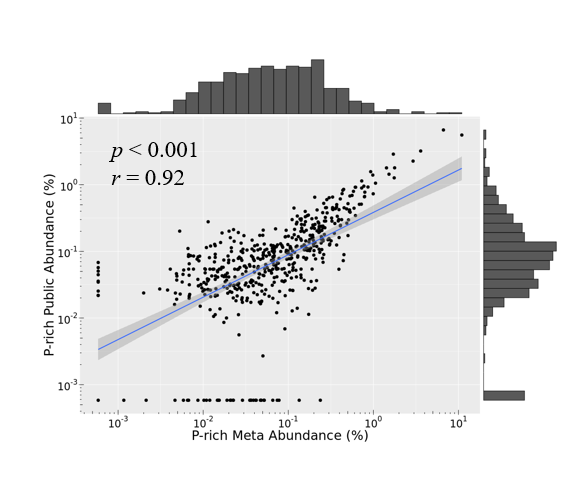
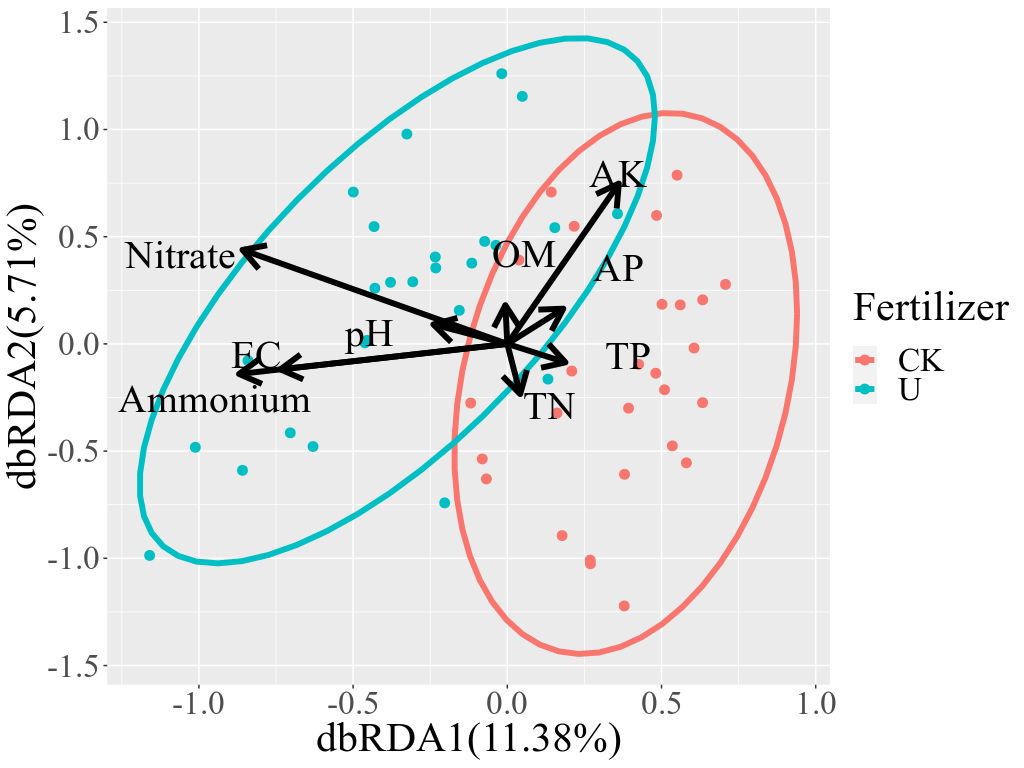
These figures above are from the two papers published by me as the first author. The R code used to analyze data and generate these figures can be found by clicking the figure links.
1 | Xiong Yi, Zheng Lu, Meng Xiangxiang, et al. Protein sequence databases generated from metagenomics and public database produced similar soil metaproteomic results of microbial taxonomic and functional changes. Pedosphere, 2021 |
SiprosToolKits-Sipros4
Sipros 4 tutorial
Shotgun metagenomic tutorial
16S amplicon tutorial
Sipros GUI
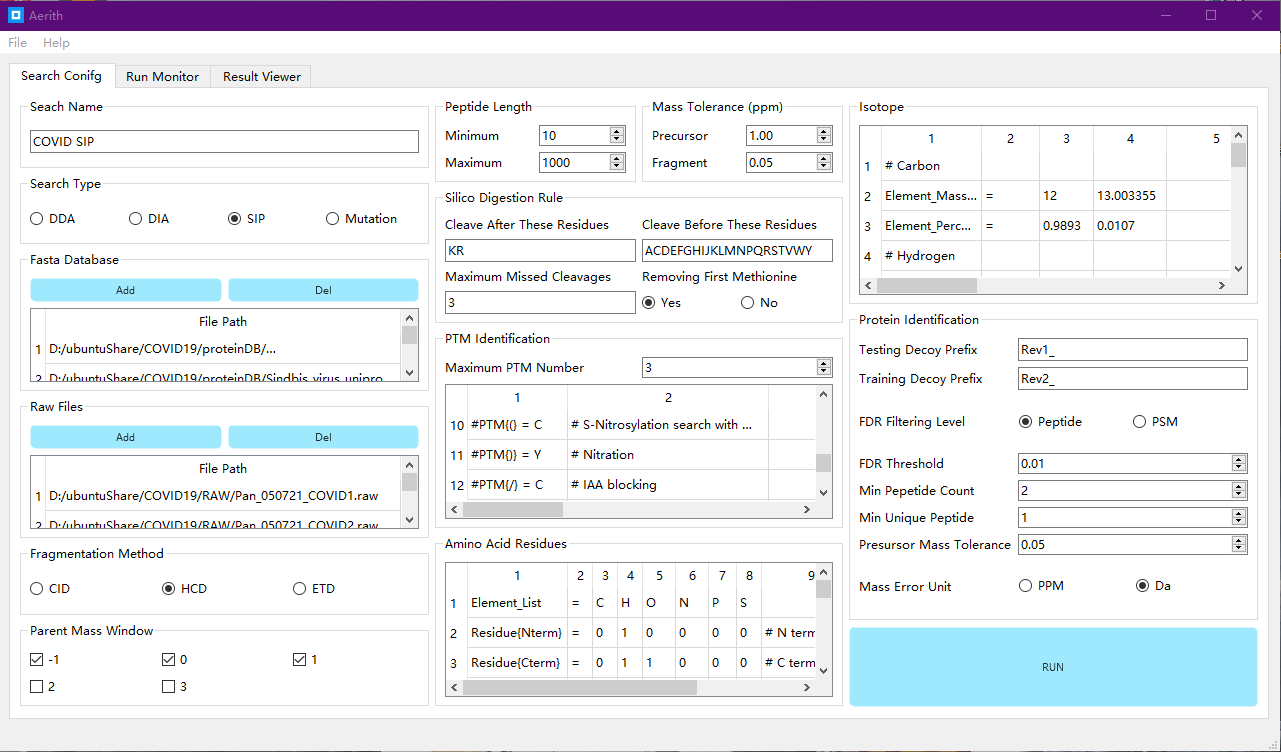
This project aims to reconstruct our command-line mass spectrum analysis tool Sipros by QT and C++. The new user interface has a user-friendly GUI. The reconstructed search engine is faster and more accurate with enhanced function in stable isotope-labeled proteomic research. The released software package is cross-platform, portable, and can be easily deployed by anaconda.
JGI Data Mining
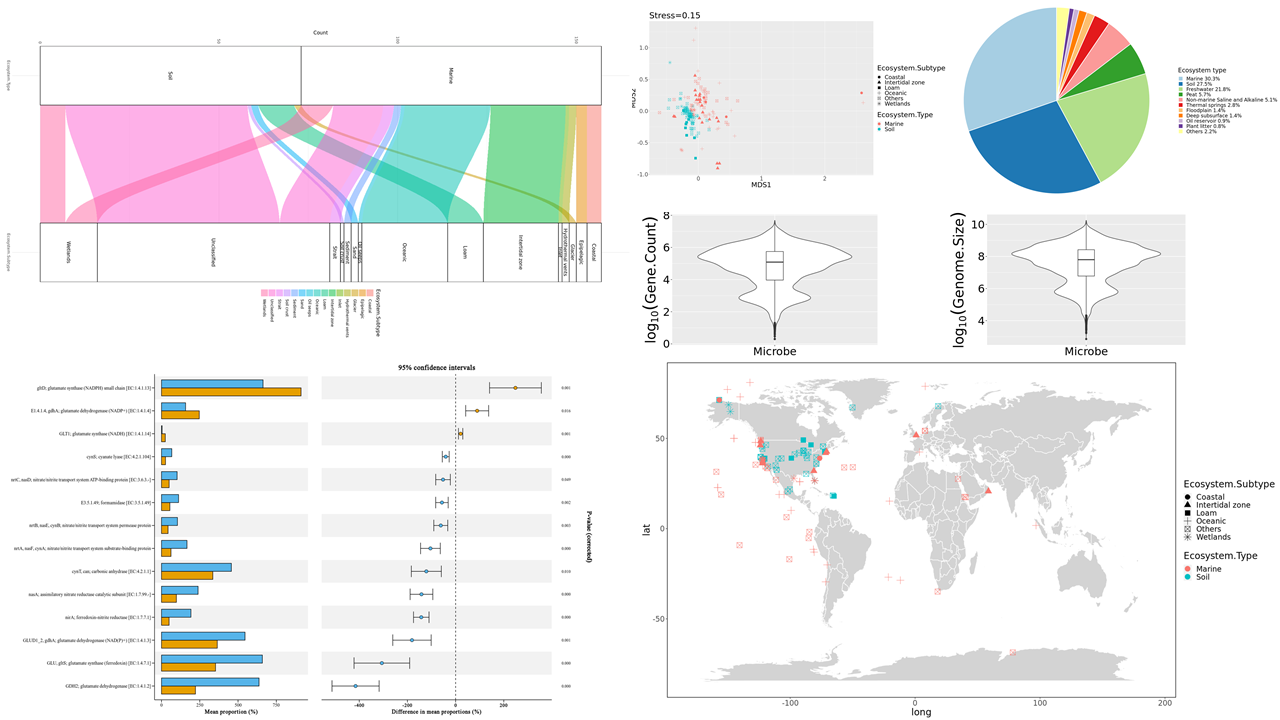
The R code used to analyze data and generate these figures can be found by clicking the title link.
The microbial nucleotide sequences database’s development made it possible to simultaneously analyze the published metagenomic data from a bulk of samples. The Integrated Microbial Genomes & Microbiomes(IMG/M) system is a platform to annotate, analyze, and distribute microbes’ genomic data stored at DOE’s Joint Genome Institute (JGI) from scientific projects all over the world. Based on ensembled metagenomic annotation results from 11404 samples with the environmental information provided by the IMG database, we analyzed the gene abundance about essential biogeochemical progress, especially the anaerobic process in different microbial taxons in various ecosystem types globally. Besides, we explored the correlation between gene abundances in different biogeochemical pathways and the correlation between gene abundance dissimilarities and geographic distance. The occurrence network of microbial taxons and gene abundances in different ecosystems were also founded.
The systematic error leads to overestimate of rare species
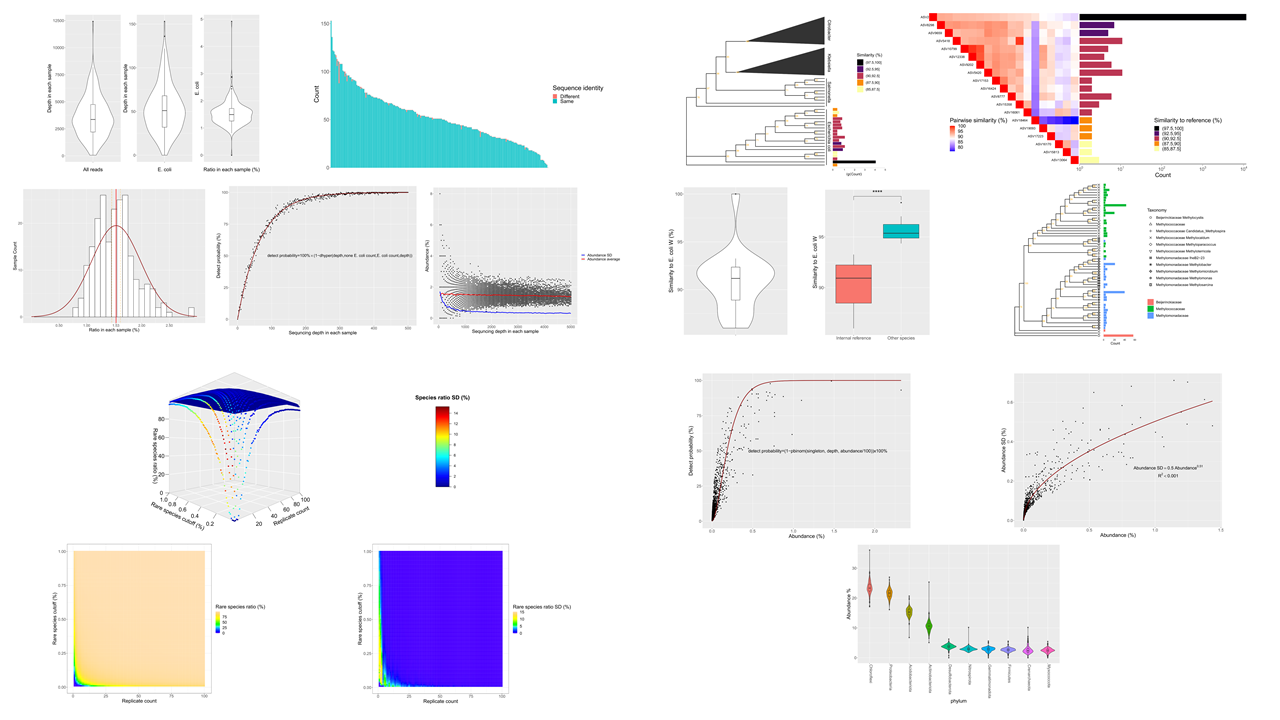
This project aims to mine one old 454 sequencing dataset to find the influence of amplicon PCR and sequencing error on rare species by DADA2 denoising and Qiime2. The R code used to analyze data and generate these figures can be found by clicking the title link.
Proteomic research of Vero cell line infected by COVID-19 virus
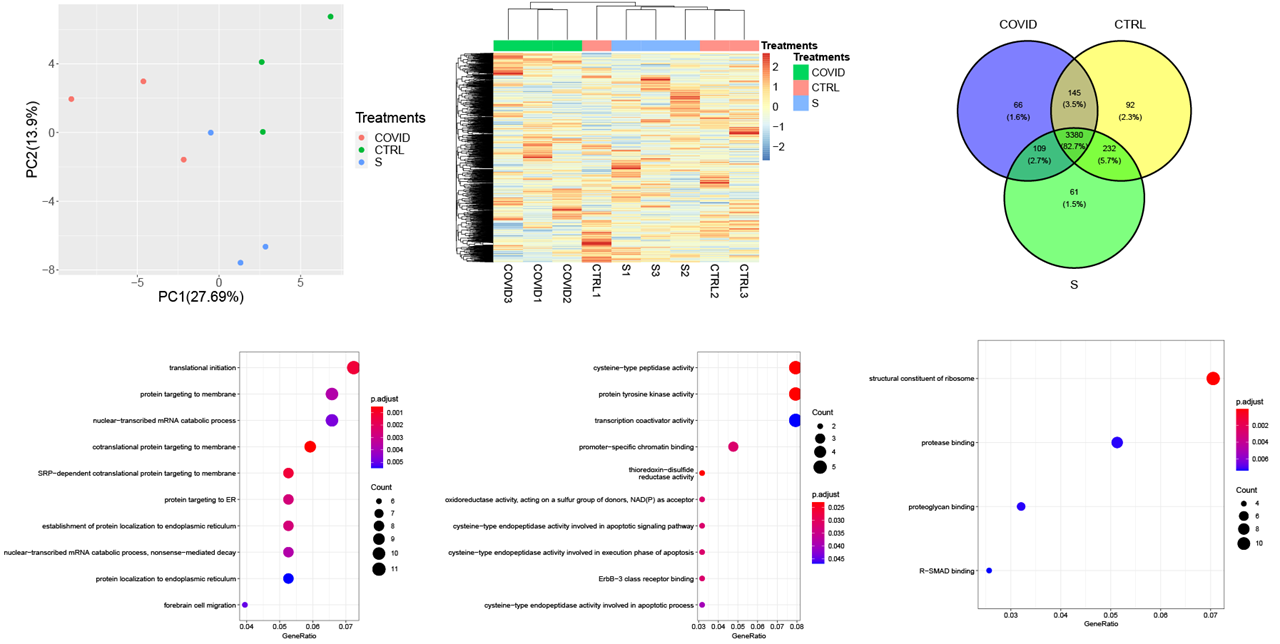
This project focuses on medical proteomic microbiology research of COVID-19 virus. We hope we can use the advanced proteomic analysis tools to decipher the pathway and regulatory network evolved in the disease progress and find the key biomarker of therapy target. The R code used to analyze data and generate these figures can be found by clicking the title link.